Our research
Our lab studies the Six family of transcription factors and the critical role its members play in myogenic differentiation. We are interested in understanding, at the molecular level, the mode of action of these transcription factors, and how they might cooperate with other transcription factors involved in skeletal myogenesis. To this end, we use functional genomic approaches such as ChIP-seq, RNA interference, expression profiling and bioinformatic analyses.
DNA binding by Six factors
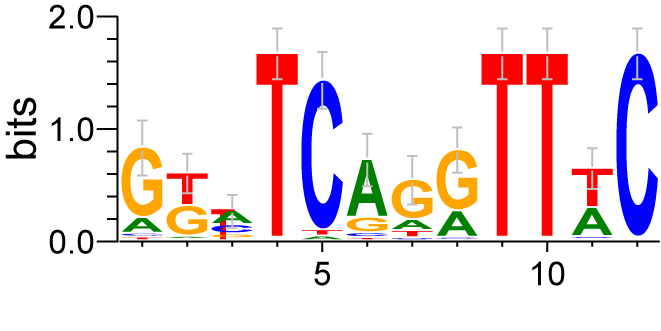
Six factors bind to DNA via their homeodomain. The DNA sequence recognized by Six1 and Six4 has been shown to conform to the MEF3 sequence motif. Using genome-wide binding profile of Six1 in myoblasts, we have determined that Six1 in fact recognizes DNA sequences related to the MEF3 motif, but that these sequences can diverge to a significant extent from the MEF3 consensus. We have identified a sequence motif that better reflects the true preference of Six1 for DNA sequences, and that enables a much better prediction of Six1 binding sites that previously reported motifs. We are pursuing this work by looking more closely at the determinants of Six1 DNA binding preferences.
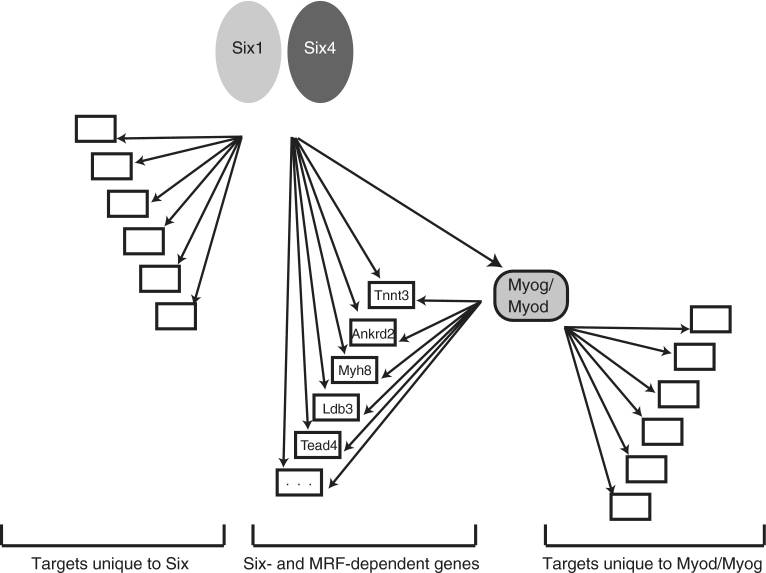
Genomic targets of Six factors
We have reported the identification of Six1 transcriptional targets in myoblasts using ChIP-on-chip. We have found that Six1 is necessary for the activation of the expression of a large number of genes, during myogenic differentiation. What is more, Six1 targets many of the same genes as MyoD: up to 40% of Six1 targets have a MyoD binding site in their vicinity. This suggests a model where Six factors cooperate with the myogenic regulatory factors to activate myogenic gene expression, during differentiation.